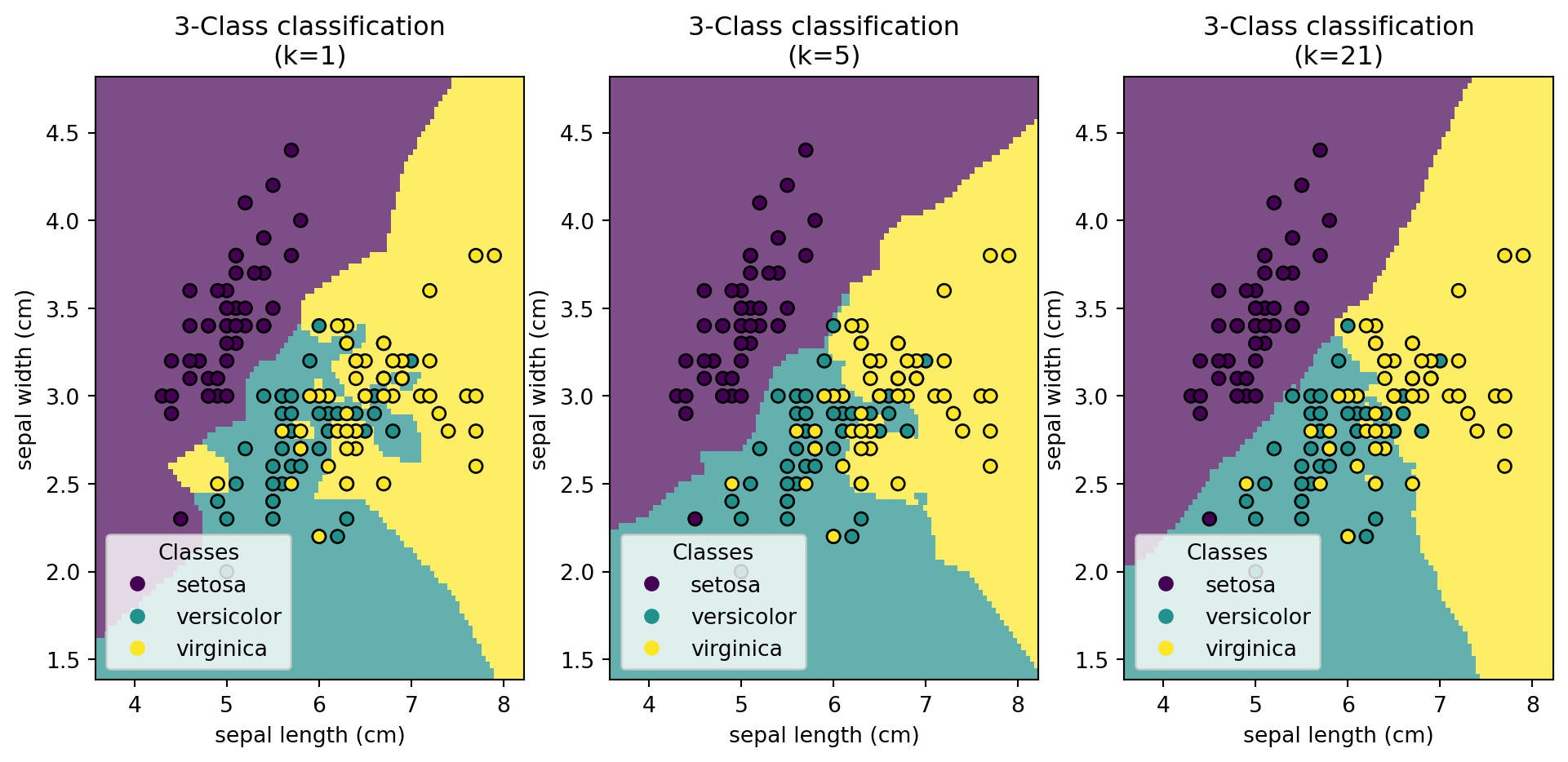
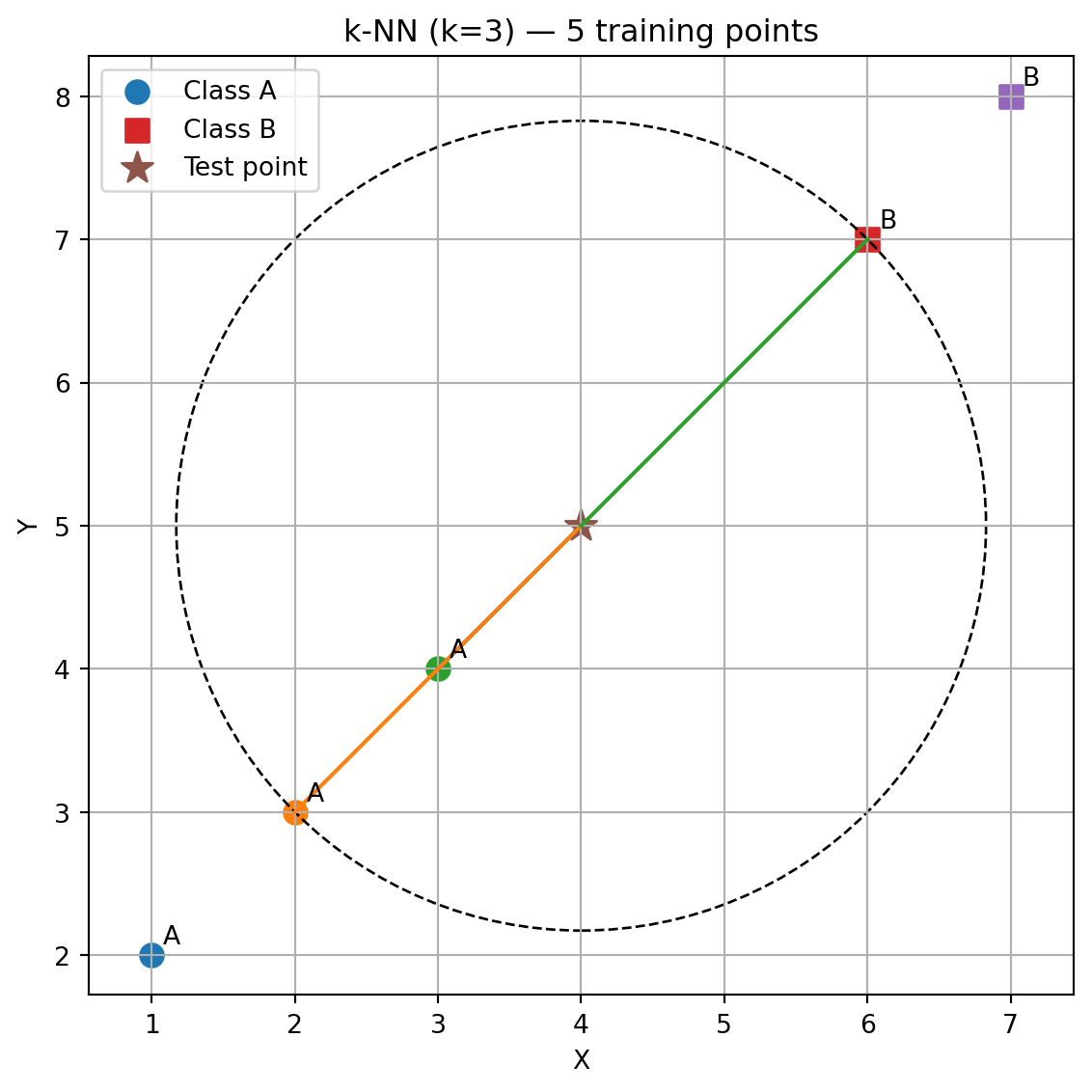
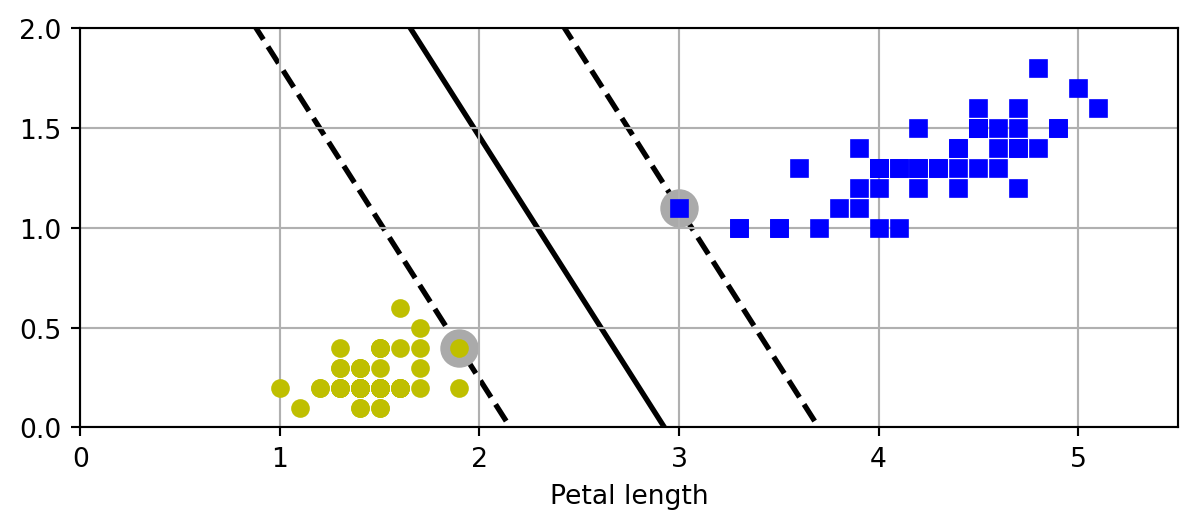
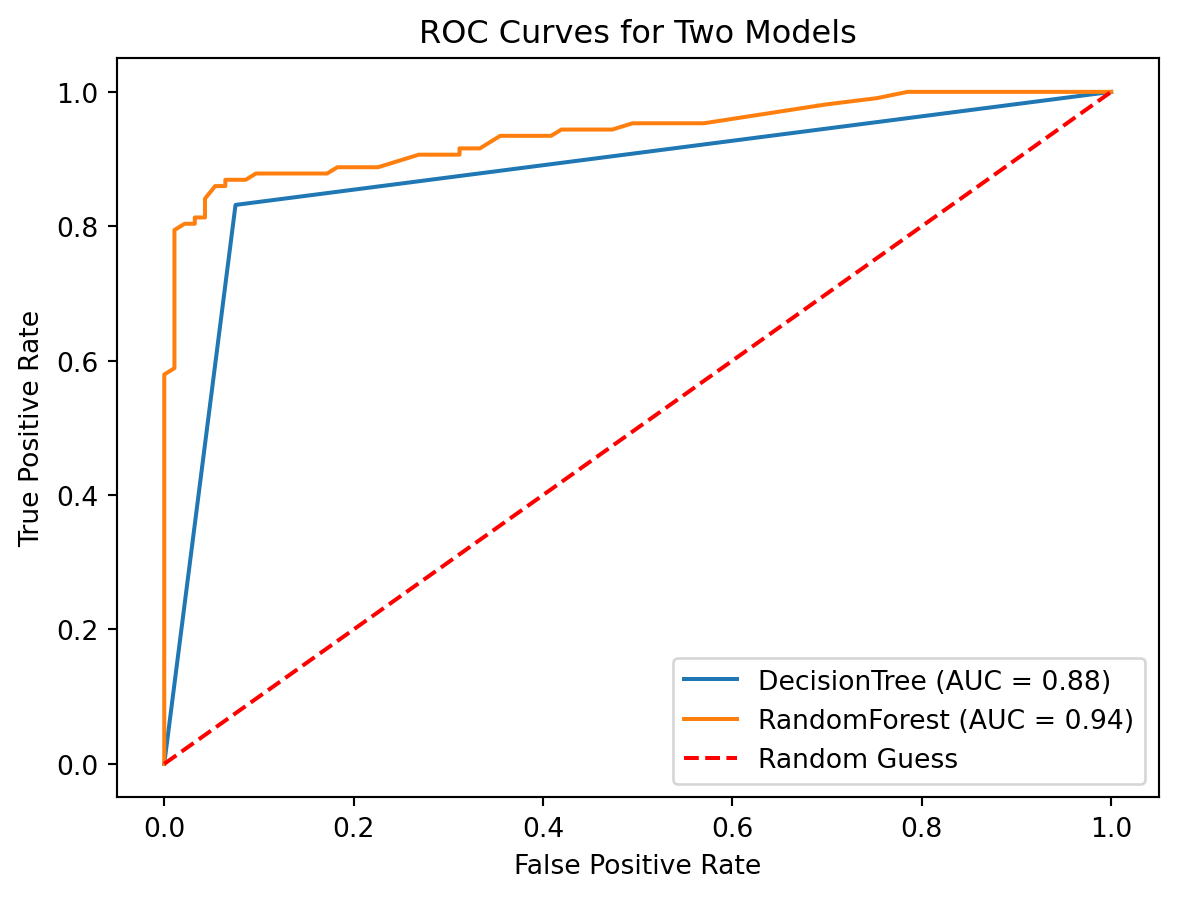
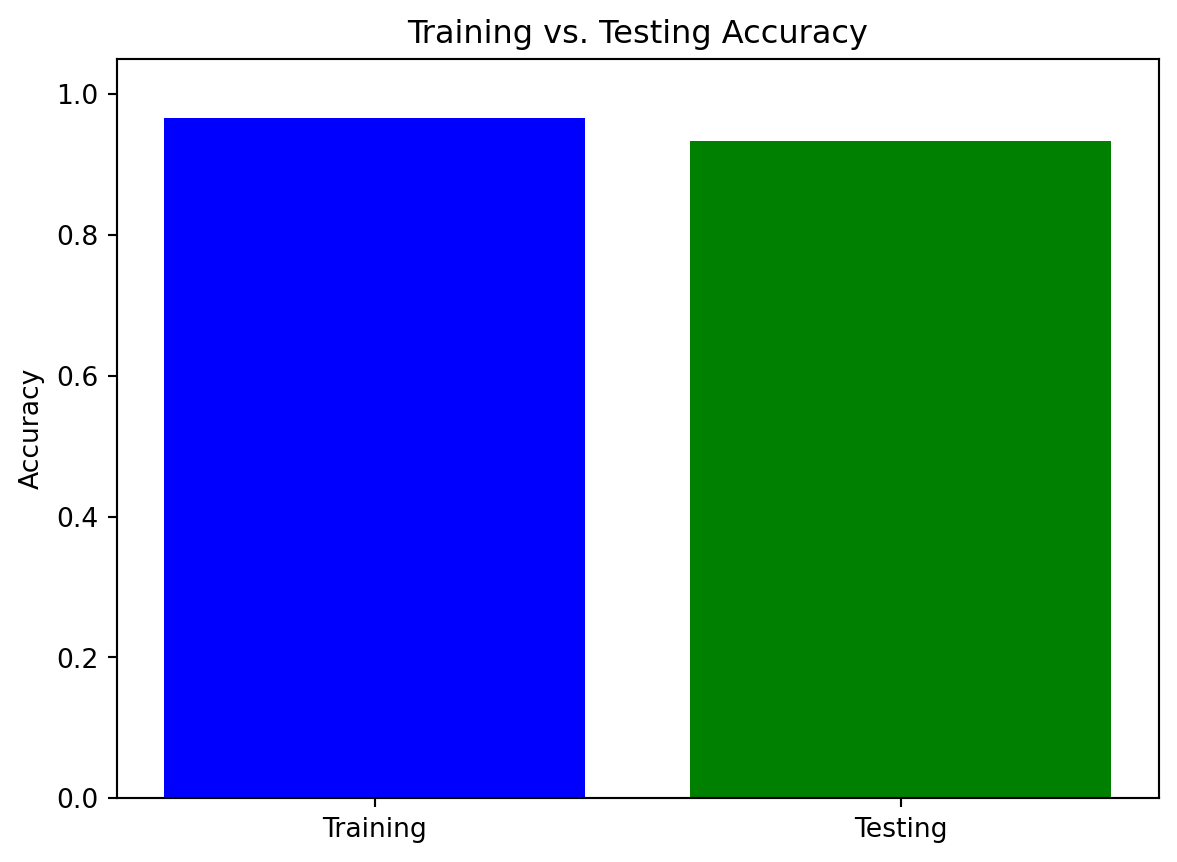
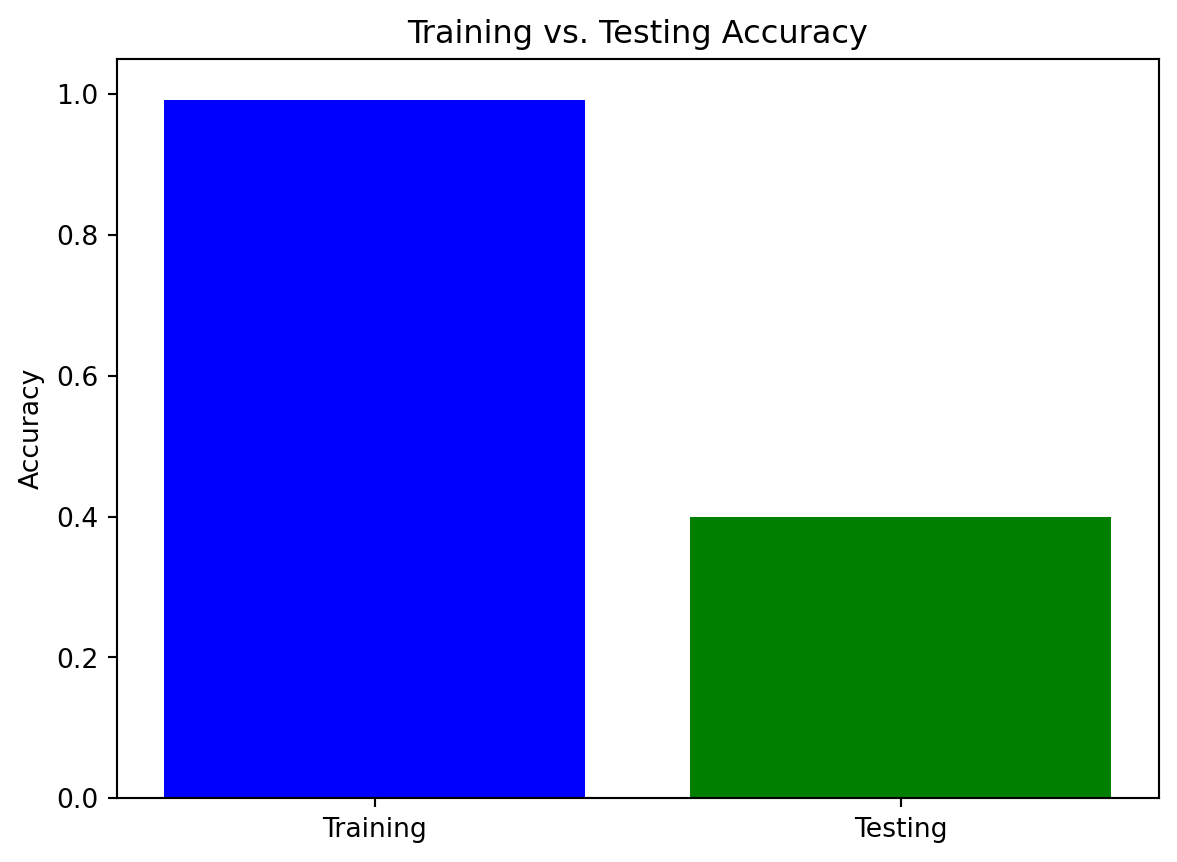
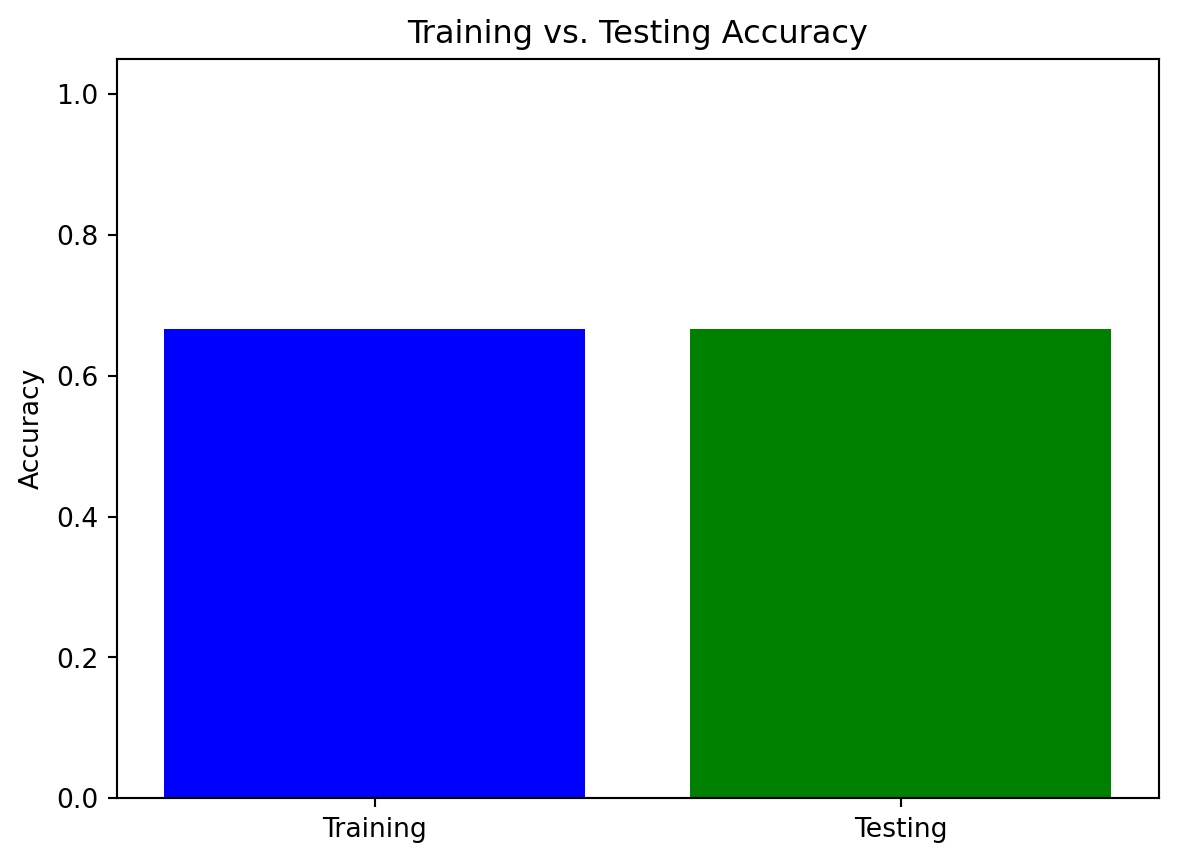
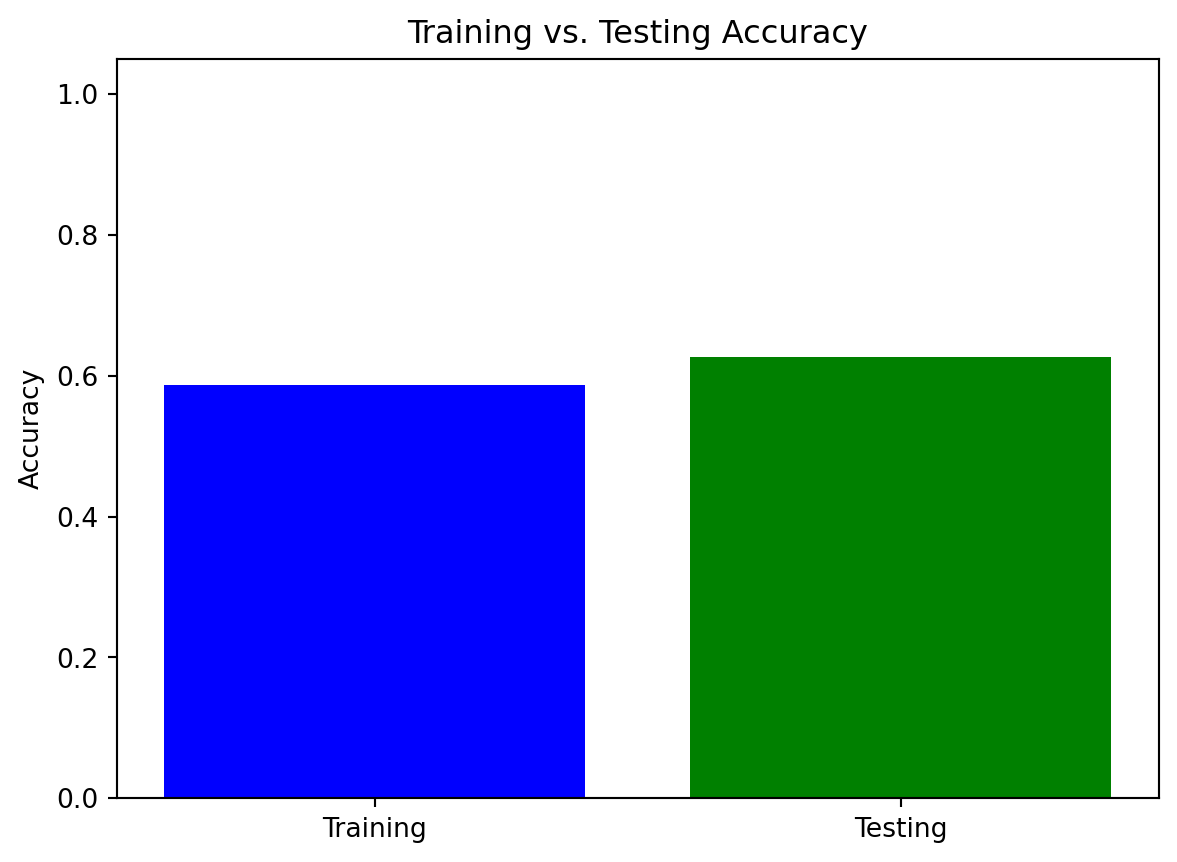
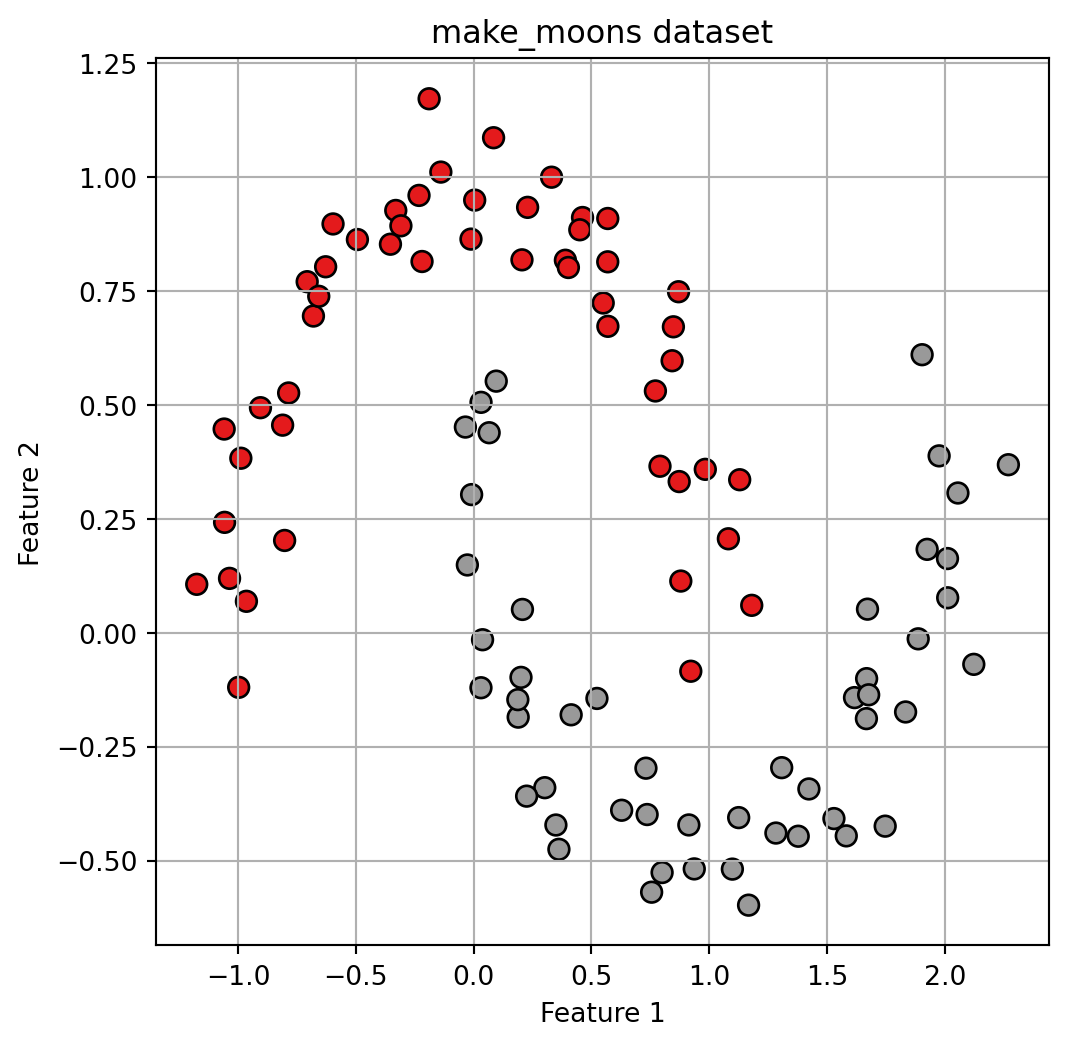
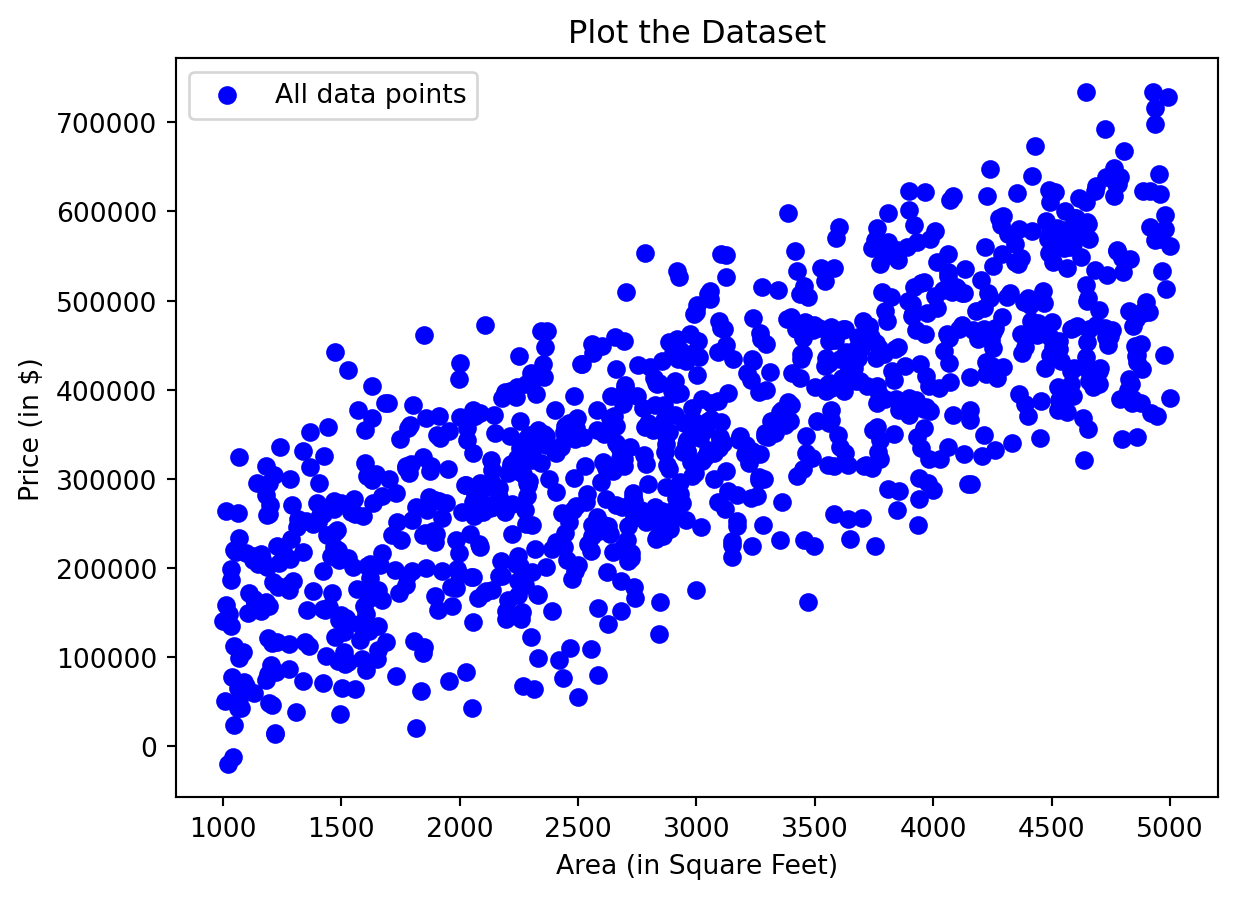
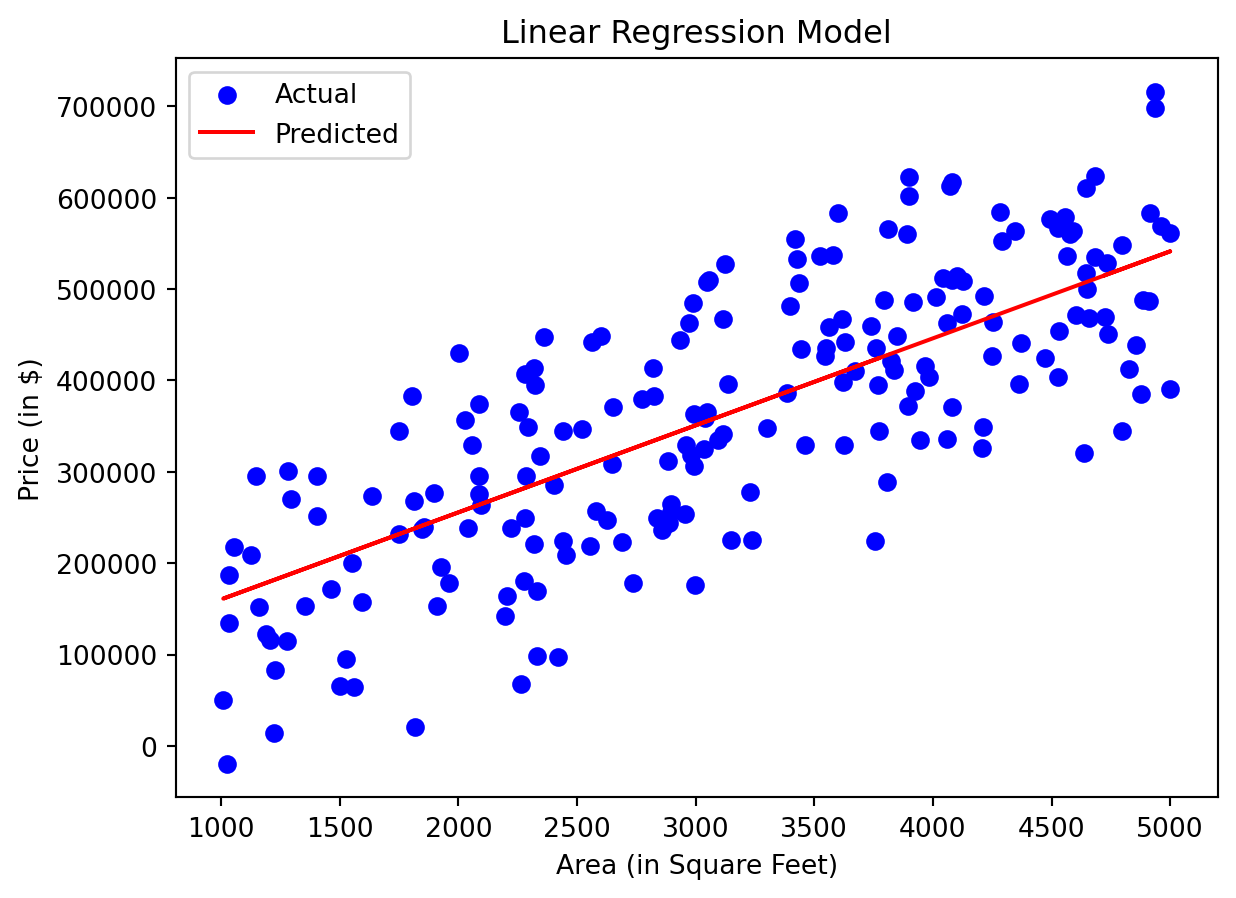
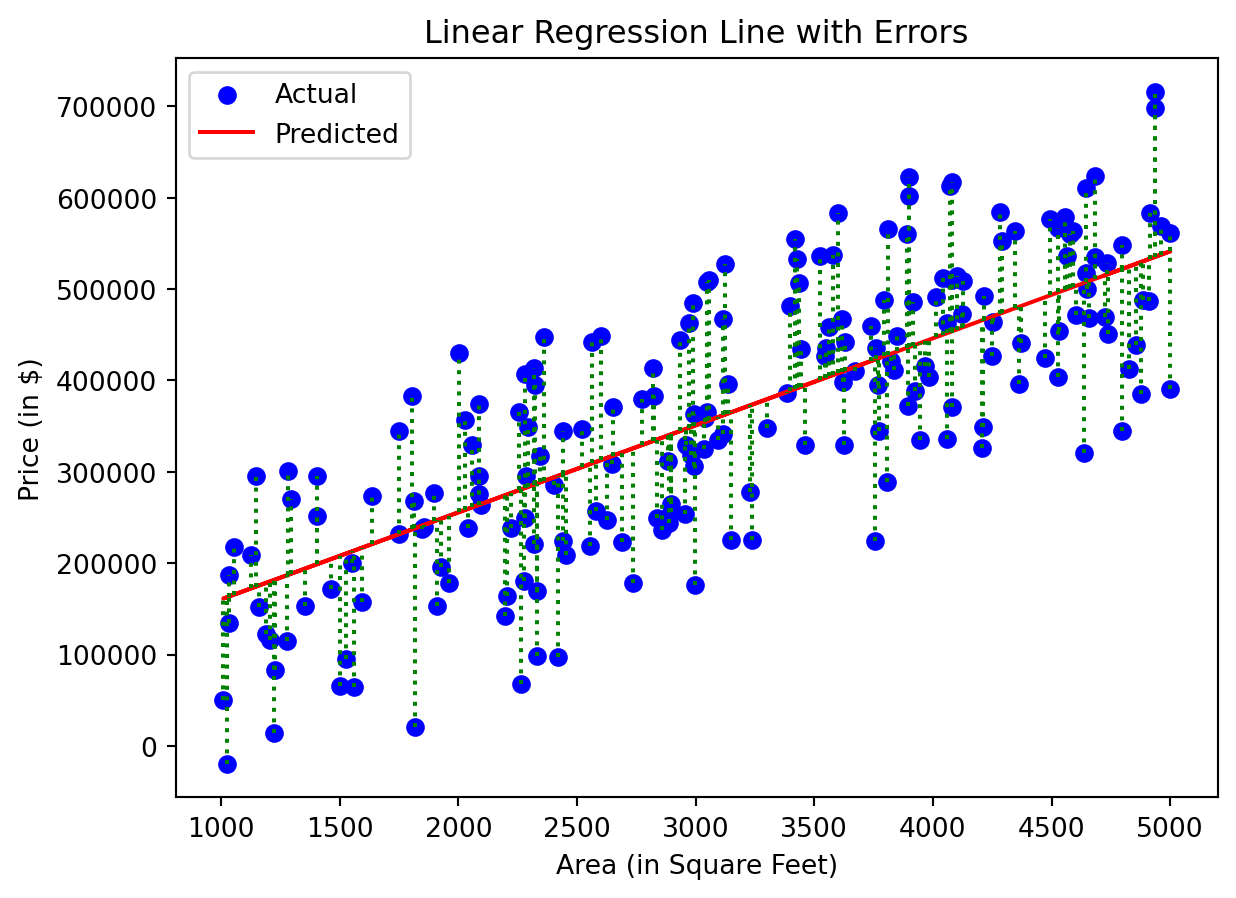
import pandas as pd
from sklearn.tree import DecisionTreeClassifier, export_text, plot_tree
import matplotlib.pyplot as plt
# Load the cat dataset mentioned in class
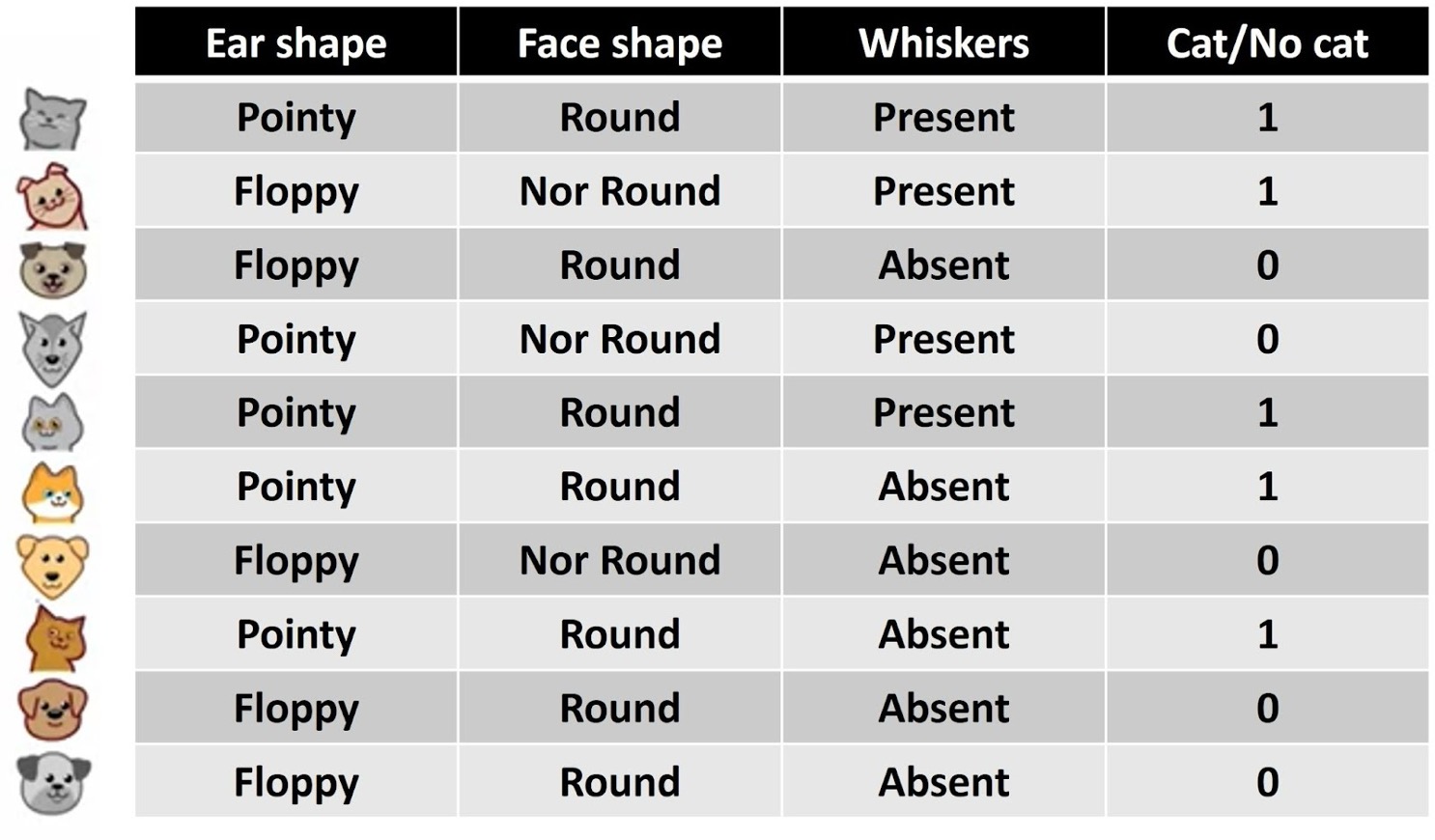
data = [
{"Ear shape":"Pointy", "Face shape":"Round", "Whiskers":"Present", "Cat/No cat":1},
{"Ear shape":"Floppy", "Face shape":"Not Round", "Whiskers":"Present", "Cat/No cat":1},
{"Ear shape":"Floppy", "Face shape":"Round", "Whiskers":"Absent", "Cat/No cat":0},
{"Ear shape":"Pointy", "Face shape":"Not Round", "Whiskers":"Present", "Cat/No cat":0},
{"Ear shape":"Pointy", "Face shape":"Round", "Whiskers":"Present", "Cat/No cat":1},
{"Ear shape":"Pointy", "Face shape":"Round", "Whiskers":"Absent", "Cat/No cat":1},
{"Ear shape":"Floppy", "Face shape":"Not Round", "Whiskers":"Absent", "Cat/No cat":0},
{"Ear shape":"Pointy", "Face shape":"Round", "Whiskers":"Absent", "Cat/No cat":1},
{"Ear shape":"Floppy", "Face shape":"Round", "Whiskers":"Absent", "Cat/No cat":0},
{"Ear shape":"Floppy", "Face shape":"Round", "Whiskers":"Absent", "Cat/No cat":0},
]
df = pd.DataFrame(data)
# Split the dataset into features (X) and target variable (y)
X = pd.get_dummies(df.drop(columns=["Cat/No cat"]))
y = df["Cat/No cat"]
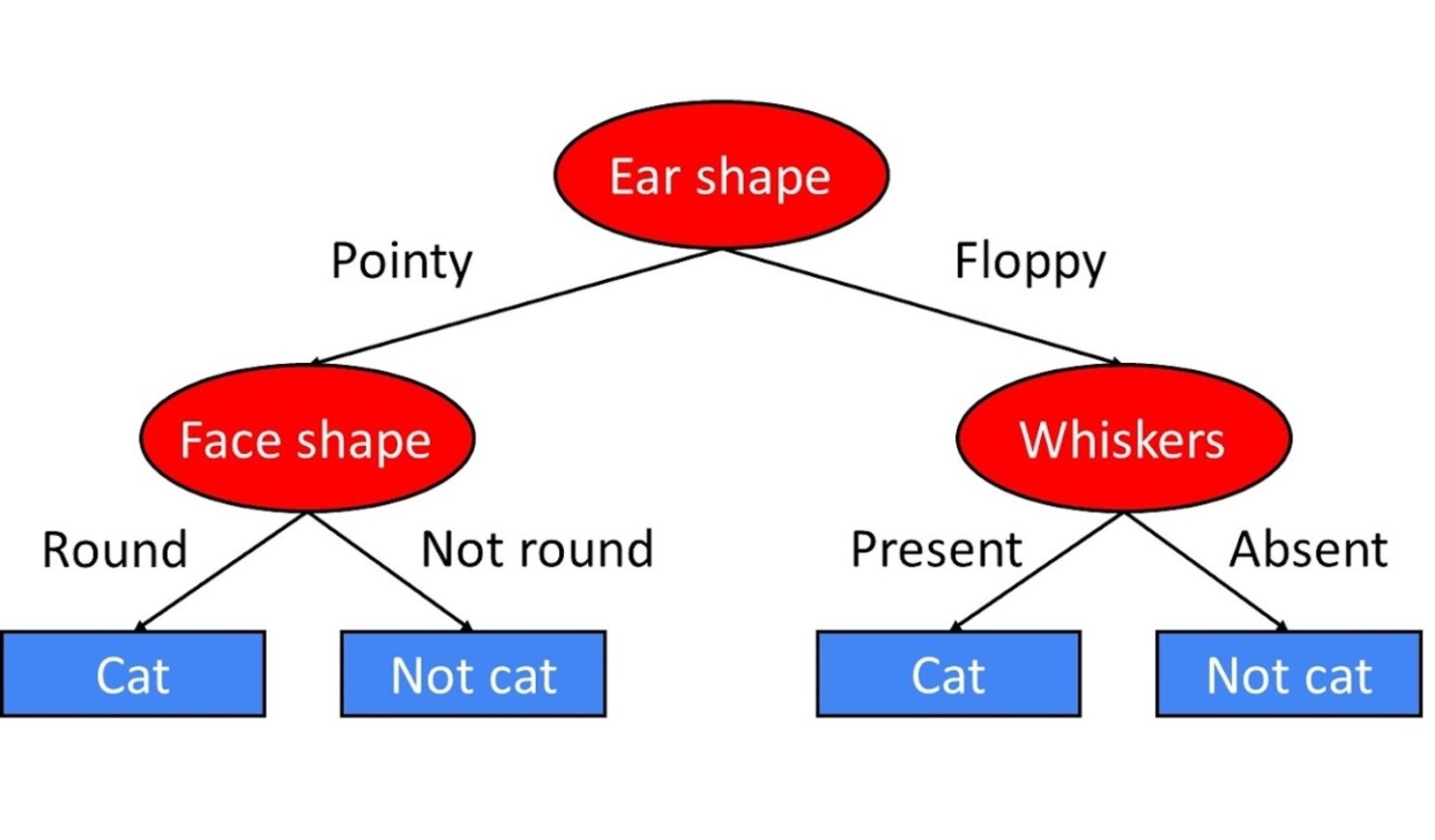
# Train our decision tree (we use entropy for IG in the sklearn library)
# Not limit the depth of the tree (max_depth=None) to allow full growth
clf = DecisionTreeClassifier(criterion="entropy", max_depth=None, random_state=0)
clf.fit(X, y)
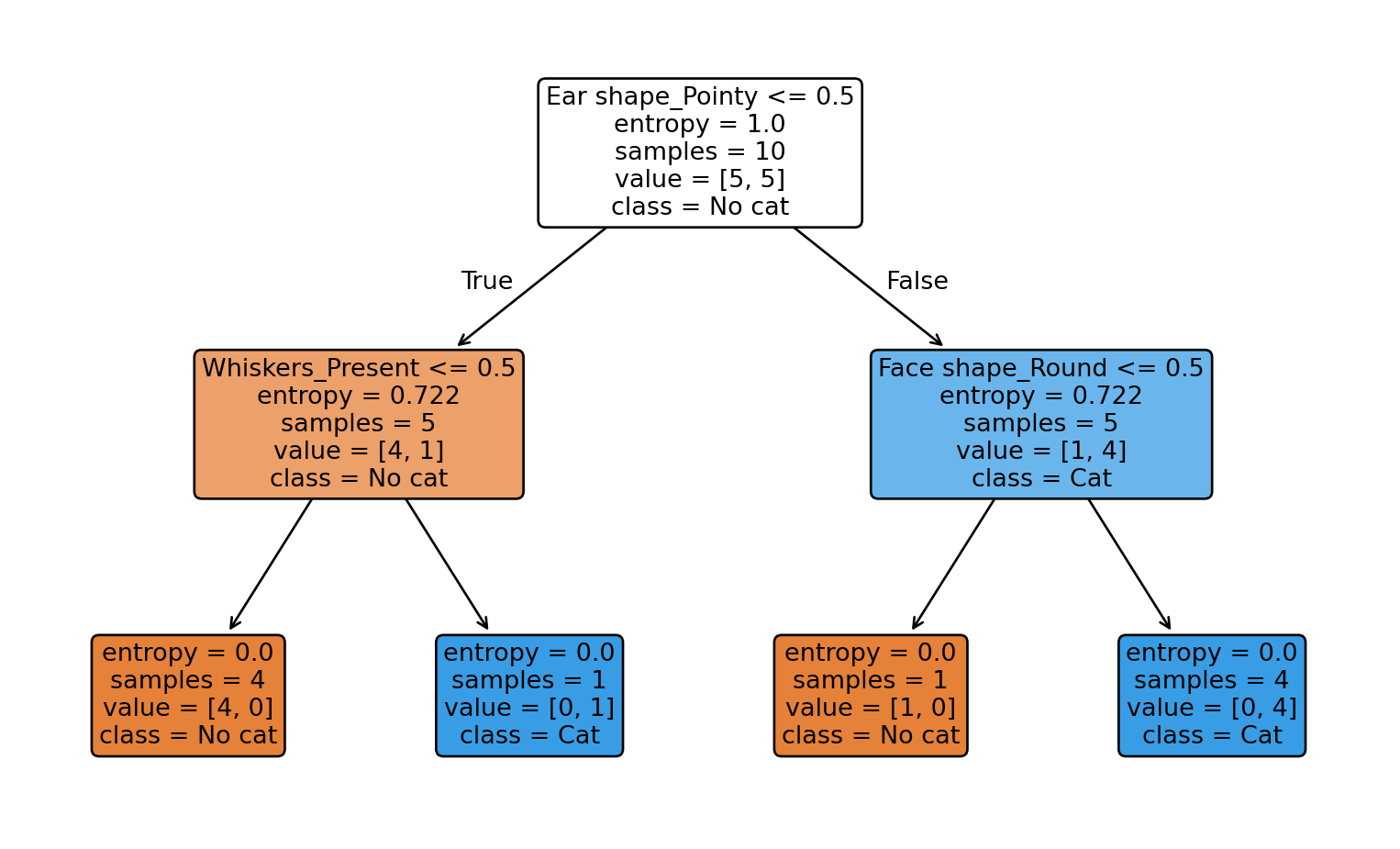
# Show decision rules
print("Decision Tree Rules:\n")
print(export_text(clf, feature_names=list(X.columns)))
# Plot the tree use the plot_tree function
print("Decision Tree Diagram:\n")
plt.figure(figsize=(10,6))
plot_tree(clf, feature_names=X.columns, class_names=["No cat","Cat"], filled=True, rounded=True)
plt.show()